Trung Do*, Phuong M. Nguyen, Phuong L. Trinh, and Yen Pham*
Abstract
Acute myeloid leukemia (AML) is one of the high mortality-rate malignant hematological diseases, hence, timely and accurate determination of prognostic factors, especially genetic abnormalities, is essential for AML therapy. Mutations of FMS-like tyrosine kinase 3(FLT3) have been indicated in holding enzyme in the active conguration, resulting in abnormal differentiation of leukocytes characterized in AML patients. The two most common FLT3 mutations, internal tandem duplications (ITD) and tyrosine kinase domain (TKD) nucleotide substitutions, are considered poor prognosis factors affecting treatment decision. Duplications within the juxtamembrane domain are found in 20-25% of cases with normal karyotype, while point mutations in the activation loop located in TKD account for 7% cases. This study established and optimized two procedures to detect FLT3 mutations in 3 main steps: (1) Amplification of FLT3 fragments with specific primers using polymerase chain reaction (PCR); (2) Identification of mutant and normal samples by electrophoresis on agarose and polyacrylamide gel (fragments containing FLT3-TKD were digested by EcoRV before electrophoresis); (3) Comparison of band pattern with the appropriate controls generated in our laboratory. Preliminary screening of 156 Vietnamese AML patients revealed 14 FLT3-ITD and 6 FLT3-TKD mutations. The electrophoresis gels showed that ITD insertions ranged from 1 to 3 in number, 20 to 90 bp in length and 4.6% to 63.7% in mutation level. The sequencing analysis denoted that 5 ITD sequencing samples including 1 in intron and 4 in exon 14, all of which did not shift the open reading frame, whilst 5 TKDs had mutant levels varied from 12.77% to 32.78% and contained G to T substitution in codon 835, resulting in Asp to Tyr amino acid change (D835Y).
Introduction
Acute Myeloid Leukemia (AML) is one of the four types of cancer affecting leukocytes, characterized by the rapid growth of abnormal white blood cells that accumulate in the bone marrow and interfere with the production of normal blood cells (1). Previously, diagnosis and treatment for AML patients were often based on clinical and cytogenetic characteristics, especially, age and karyotype, however, patients with normal karyotype (AML-NK) responded differently to the same treatment regimen (2,3). Thus, further researches were conducted at themolecular level and various gene mutations were detected and divided into two groups: one group included mutations that activate signaling pathways, increasing proliferation and accumulation of blood cells in the marrow born; the other contained mutations that affect transcription factors or components ofcell cycles, leading to a reduction of cellular differentiation (2,4). Among AML-linked gene mutations, those occuring on Nucleophosmin 1 (NPM1), FMS-like tyrosine kinase 3 (FLT3) and CCAAT/enhancer binding protein α (C/EBPα) were most frequent with prevalences of 45-64%, 28-34%, 10-18% in AML-NK (respectively) (3). It is important to note that in these 3 genes,occurance of FLT3 mutations of AML was associated with poor prognosis and detection of significantly change treatment towards FLT3 inhibitor (4-12). In 2001, Whitman reported that 23/82 AML patients with FLT3-ITD had disease-free survival rate after applied a treatment lower than those with normal FLT3(FLT3-WT) (4).
FLT3 is a class III receptor tyrosine kinase (RTKIII) expressed in hematopoietic progenitor cells(13-16). The two most commonFLT3 mutations include internal tandem duplications (FLT3-ITD)of the juxtamembrane (JM) domain, found in 20% - 30% of patients with AML-normal karyotyping (NK), andpoint mutation of tyrosine kinase domain (FLT3-TKD),diagnosed in 7% AML-NK cases (5, 17). These mutations alter the configuration of the constitutive activation, resulting in increased access ofsubstrates and ATPthat leads to autophosphorylation and subsequently triggers various intracellular signaling pathways (16-20). Presence of FLT3 mutations were reported to cause protective FLT3-mutation cells, leading todrug resistance to FLT3 inhibitors and cytotoxic agents (21-23). Thus, identification and characteration of FLT3 mutations are essential to point out therapeutic strategies for AML patients (24).
FLT3-ITD involves a segment of the JM domain-coding sequence (exons 14 and 15) causes ligand-independent receptor dimerization and constitutive activation of the TK domains, leading to activate signaling pathways (6, 19, 20, 25). Most of FLT3-ITD mutations were from 6 to 180 bp in length (median, 54 bp), thus could be detected by polymerase chain reaction (PCR) combined with agarose gel electrophoresis (26, 27).
FLT3-TKD mutations are point mutations in the activation loop of FLT3 gene, usually occurring in codon D835, and less frequently in codon I836 (5, 29). Most often, these result in replacement of aspartate at the 835th posititon by another amino acid (e.g. tyrosine, histidine, valine, or glutamate) (5, 30). PCR-RFLP (restriction fragment length polymorphism) using EcoRV is a regular detection method for FLT3-TKD. It is important to note that digestion by EcoRV could not detect all FLT3-TKD mutations (5, 7), however, sequence analysis showed that 50 – 68% of FLT3-TKD mutations were substitutions of the first nucleotide of codon D835, most commonly from G to T (5, 29, 30).
In this study, we used two established diagnosis procedures to detectFLT3-ITDand FLT3-TKD mutations and preliminarily characterized those in Vietnamese AML patients through amplification ofFLT3 gene, gel electrophoresis and mutant sequencing with EcoRV digestion as an additional step for FLT3-TKD mutation before electrophoresis.
Materials and Methods
Sample collection and DNA isolation
Total blood samples from 156 AML patients diagnosed by karyotyping and cellular morphological-chemical techniques were provided by the National Institute of Hematology and Blood Transfusion. Genomic DNA was isolated by QIAamp DNA blood mini kit, then measured for quality and quantity by electrophoresis on 1% agarose gel and Nanodrop 2000c. Samples which were suitable for further PCR amplification had DNA concentrations between 20-400 ng/µl and A260/A280 values from 1.8 to 2.
PCR amplification of FLT3-ITD fragments
The sequences of oligoprimers used to amplify the target 329 bp DNA fragment from exon 14-15 of FLT3 gene were: FLT3_forward (5’-GCAATTTAGGTATGAAAGCCAGC-3’) and FLT3_reverse (5’-CTTTCAGCATTTTGACGGCAACC-3’) (19) (IDT). The 25 µl PCR reaction was mixed 20 ng of genomic DNA; 10 pmol of each primer; 0.2 mM for each deoxyribonucleotide triphophate; 1 unit of Dream Taq DNA polymerase (Thermo Scientific) and 2.5 µl of 10X Dream Taq Buffer and performed with the protocol: 94°C for 5 minutes; 35 cycles of 94°C for 1 minute, 56 °C for 45 seconds, 72°C for 1 minute; and finally 72°C for 10 minutes. The resulting PCR products were analysed by electrophoresis on 2% SEAkem and 3% Ultraphor agarose. In addition, an amount of 2 µl of the PCR products was run in the capillaries of the Advanced Analytical Fragment Analyzer system. The results were analyzed using ProSize 2.0 software which produced digitalized gel images.
PCR amplification of FLT3-TKD fragments
D835 and I836 are encoded by the nucleotides GATATC, which form the EcoRV recognition and restriction site (12). 114 bp-fragment containing this sequence was amplified from exon 17 of the FLT3 gene using the primers FLT3-TKD forward (17F: 5’-CCGCCAGGAACGTGCTTG-3’) and FLT3-TKD reverse (17R: 5’-GCAGCCTCACATTGCCCC-3’) (5) (IDT). PCR was performed with a total volume of 25 µl containing 50 ng of genomic DNA, 2 pmol of each primer, 0.2 mM for each deoxyribonucleotide triphophate, 1 unit of Dream Taq DNA polymerase (Thermo Scientific), 2.5 µl of 10X Dream Taq Buffer and 0.875 µl DMSO. The thermal cycle used was: 94°C for 5 minutes; 30 cycles of 94°C for 30 seconds, 58 °C for 30 seconds, 72°C for 30 seconds; and finally 72°C for 10 minutes. PCR products were analyzed by electrophoresis on 12 % gel polyacrylamide and ImageJ software was used to estimate PCR product concentrations from gel images.
Digestion of PCR products by EcoRV
For EcoRV digestion, 6-8 ng of PCR product were added directly to a mixture containing 2u enzyme (New England Biolabs), 1 µl of restriction buffer (10x buffer B: 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 100 μg/ml BSA, pH 7.9 at 25°C), and DNA free water up to 10 µl. The reaction mixture was incubated for 30 minutes at 37°C, then inactivated at 80oC for 20 minutes. The digestion products were analyzed by 12% polyacrylamide gel electrophoresis. Theoretically, normal samples produce two digested fragments of 68 and 46 bp after treated with EcoRV while PCR products containing D835/I836 mutations remain intact (5).
Determination of mutant sequences
Extra band of 329 bp-products or EcoRV-undigested fragments were cut out from the agarose gel, purified and cloned into pGEM T easy vector (Promega). The vector was transformed into Escherichia coli DH5α competent cells. Screening of transformants was performed by white-blue screening method. Verification of positive colonies were done by PCR with vector primers: the pGEM T easy forward (5’-GCTGCAAGGCGATTAAGTTG-3’) and the pGEM T easy reverse (5’-GTTGTGTGGAATTGTCACG-3’), or following by EcoRV digestion. Finally, plasmid DNAs purified from the screened transformants were sent for sequencing.
Results and Discussions
Diagnostic procedure of FLT3-ITD and FLT3-TKD mutations
Detection of FLT3-ITD mutations through PCR combined with gel electrophoresis
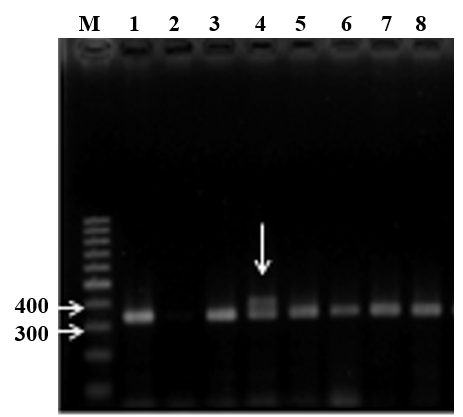
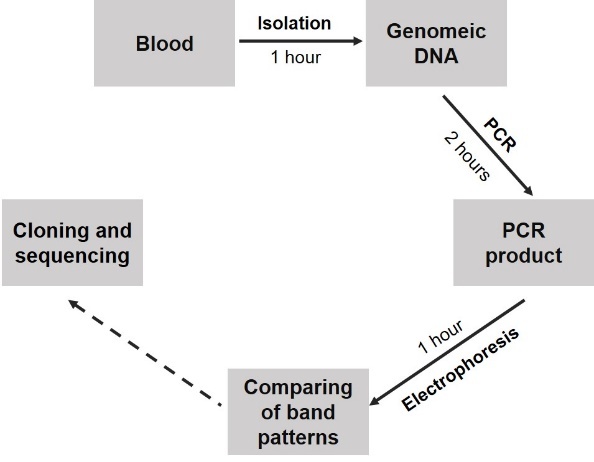
To identify FLT3-ITD, the PCR was used to amplify a 329 bp fragment in exon 14-15 from isolated genomic DNA optimized thermal conditions and amount of components shown as a single DNA band between 300-bp and 400-bp bands of the DNA marker (Figure 1A). In some samples (Figure 1A, lane 4), the electrophoresis image showed extra bands at higher molecular weight positions in addition to the wild type fragment. Sequencing results validated that these band corresponded to duplications, thus we concluded that PCR protocol followed by electrophoresis was efficient for detection of the ITD mutations in FLT3. Finally, the step by step diagnostic procedure was briefed in Figure 2A with sensitivity from approximate 104 copies.
 |
 |
| A | B |
|
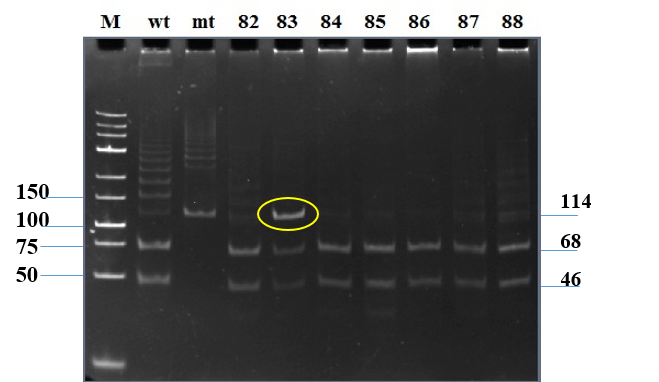
Figure 1. Detection of the FLT3 mutations through PCR combined with electrophoresis analysis. A - Separation of FLT3-ITD mutations on 2% agarose gel. M: 100 bp DNA marker; lane 1: a normal sample; lane 2: a no-DNA template control; lanes 3-8: PCR products of AML samples. B - The electrophoresis of FLT3-TKD digested products.M: lowrange DNA marker; mt: mutant sample; wt: wild type sample; lane 82-88: digested products of 114 bp-fragment. |
|
Optimizing a PCR-RFLP procedure for FLT3-TKD mutation detection
To optimize PCR amplification, a series of annealing temperatures (from 55oC to 59oC with intervals of 1oC) combined with different DMSO concentrations (2%, 3.5%, 5%), together with primer and template concentration were applied. The result showed that the use of 58oC in annealing step and 3.5% DMSO generated a specific band migrated as a single band between 100-bp and 150-bp bands of DNA marker on electrophoresis gels (Figure 1B, lane 3-mt) which as confirmed by sequencing to have wild type sequence. Conclusively, the established PCR conditions successfully amplified the 114-bp target. The sequenced plasmid containing wild type FLT3-TKD fragment was used as normal control in further experiments.
Incomplete digestion might lead to the prevalence of the wild-type 114-bp product, resulting in false positive, thus EcoRV digestion was conducted at different enzyme: substrate ratios and digestion time using a synthetic mutant (see Materials and Methods). FLT3-TKD mutation contained G-T substitution corresponding to the first nucleotide in EcoRV recognition sequence, reported to the most frequent point mutation (5). Substrate concentration fixed between 6-8 ng was used and EcoRV amount was varied from 0.1 to 1 microliter. The reaction mixture was incubated for different times, ranging from 30 seconds to 3 hours. The experiments showed that EcoRV digestion using 0.2 microliter of enzyme was max 91.2% complete after 30 minutes.
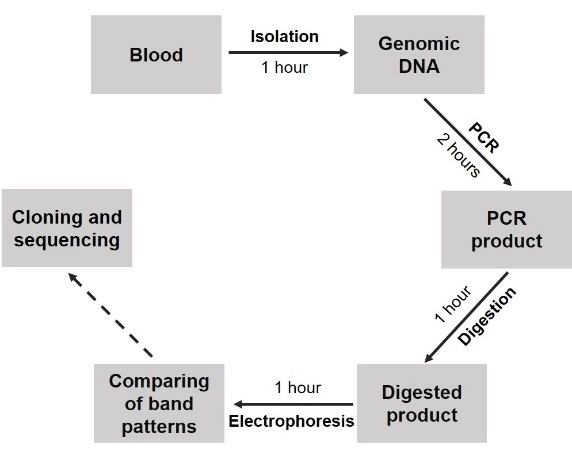
After optimization, for further experiments, the 114-bp TKD fragment was amplified with 58oC in annealing temperature and 3.5% DMSO, then was digested with 0.2 microliter of EcoRV for each 6-8 ng of the PCR product at 37oC in 30 minutes. The procedure for detection of FLT3-TKD with the sensitivity from 5% mutant DNA was summarized in Figure 2B. The total time for these procedure 4 hours and 5 hours with ITD procedure and TKD procedure, respectively, thus those were suitable for regular testing in hospital.
 |
 |
| A | B |
|
Figure 2. Diagnostic procedures for detection of FLT3 mutations. A. FLT3-ITD diagnostic procedure; B. FLT3-TKD diagnostic procedure |
|
Preliminary screening of FLT3 mutations in AML samples from Vietnamese patients
After preliminary screening of 156 Vietnamese AML patients, we found 14 samples (9.0%) and 6 samples (3.8%) containing suspected ITD and TKD mutations, respectively, in which, five samples of each mutant type were affirmed by subsequent cloning and sequencing (Table 1). ITD sequencing verified duplications at different positions within the 329-bp DNA fragment in these samples, while all of TKD mutations were nucleotide substitutions. Compared with previous researches, the frequencies of ITD (9.0%) and TKD (3.8%) in this study were similar to those reported elsewhere with 10%-15% and 7%, respectively (5, 17, 25).
Table 1: Statistics of FLT3 mutations.
|
Type of mutation
|
Number of mutant
|
Percentage (%)
|
Confirmed by sequencing
|
|
ITD
|
14
|
9.0
|
5
|
|
TKD
|
6
|
3.8
|
5
|
Molecular characteristics of FLT3 mutations
FLT3-ITD mutations
The electrophoresis analysis showed some characteristics of the mutations which might affect prognosis and treatment decision. Previous studies defined mutation level as the number of white blood cells carrying mutant alleles compared to the total number of leukocytes in percentage. In this study, the mutation level could be roughly determined based on density calculation of wild-type band and mutant band using ImageJ software. Study of 156 AML patients, mutation level was estimated to range from 4.6% to 63.7% (mutant/wild-type (wt) ratio ranged from 0.05 to 1.75) which was lower than reported in another study with ratio ranging from 0.03 to 32.56 (median, 0.78) (27).
Table 2: Summary of detected extra bands and estimated mutation levels.
|
Number
|
Insert length
(estimated, bp)
|
Insert length
(sequenced, bp)
|
Number of insertions (estimated)
|
|
1
|
35
|
33
|
1
|
|
2
|
76
|
|
1
|
|
3
|
56; 80
|
|
2
|
|
4
|
53
|
51
|
1
|
|
5
|
65
|
63
|
1
|
|
6
|
63
|
|
1
|
|
7
|
25; 59
|
24
|
2
|
|
8
|
39
|
|
1
|
|
9
|
61; 114
|
60
|
2
|
|
10
|
27
|
|
1
|
|
11
|
62
|
|
1
|
|
12
|
36; 55; 70
|
|
3
|
|
13
|
48; 76
|
45
|
2
|
|
14
|
40; 80; 96
|
|
3
|
Plotting migration marker bands and the PCR products on electrophoresis gel, allowed prediction of the duplication lengths (25-96 bp according to linear regression equation from semi-log plot). As shown in Table 2, the sequencing result identified the mutations with lengths from 24-63 nucleotide long and strongly supported the estimation based on migration distance on agarose. In a study conducted by Meshinchi and colleagues, 15-174 bp size range was reported in 52% FLT3-ITD mutation (25, 26). Clinical research demonstrated that mutation length plays an important role in the prognosis, as longer mutations affecting multiple functional components of the FLT3 protein. In relapse-free survival at 4 years from complete remission for those with shorter and longer ITDs was 51% and 19%, respectively (P = 0.035) (26). Hence, patients with longer ITDs had a worse relapse-free survival rate (8, 25, 26, 27).
In addition, electrophoresis separation showed that some samples had one extra band (larger than 329 bp), while others had 2 or more. 8/14 FLT3-ITD-positve samples had single duplication, 4/14 samples had 2 and 3 samples had 3 (Table 2).
Table 3: Sequences of five FLT3-ITD mutations.
|
Number
|
Duplication sequence: DNA (5’-3’) [Peptide (N-C)]
|
Size
|
Site (aa)
|
|
1
|
CTGATTTCAGAGAATATGAATATGATCTCAAATGGGAG
TTTCCAAGAGAAA [TDFREYEYDLKWEFPRE]
|
51 bp [17 aa]
|
608
|
|
2
|
TTCTCCTCAGATAATGAGTACTTCTACGTTGATTTCAGAGAA
TATGAATATGATCTCAAATGG [FSSDNEYFYVDFREYEYDLKW]
|
63 bp [21 aa]
|
603
|
|
3
|
CTACGTTGATTTCAGAGAATATGAA [DYVDFREYE]
|
27 bp [9 aa]
|
597
|
|
4
|
GTCAGAGAATATGAATATGATCTCAAATGGGAGTTTCCAAGAG
AAAATTTAGAGTTTGGT
|
60 bp
|
intron
|
|
5
|
TGAGTACTTCTACGTTGATTTCAGAGAATATGAATATGATCCCAA
[NEYFYVDFREYEYDP]
|
45 bp [15 aa]
|
601
|
The sequencing analysis of five FLT3-ITD mutations also indicated that duplications were directly inserted next to the original fragments and only one sample had the duplication in the intron, while all of the remaining samples had their duplications in exon 14 (Table 3). The converted acid amine sequence from the mutant nucleotide sequence showed that the reported FLT3-ITD mutations did not shift the open reading frame and were at different locations around positions from 597-608.
FLT3-TKD mutations
Sequence analysis showed that missense D835 mutations were present in all samples. The first nucleotide of D835, G was substituted with T, resulting in an Asp to Tyr amino acid change (D835Y) (Figure 3). This type of substitution was the most frequent TKD mutation followed by D835V, D835H, D835E, D835N and I836L+D (5). In this study, mutant level of FLT3-TKD was quantified based on the percentage of the remaining 114 bp fragment which was converted to mutant level by Excel Microsoft, to vary from 10.77 % to 32.78 % (Table 4). In recent researches, patients with FLT3-TKD still achieved complete remission and had overall survival and disease-free survival rate with no significant difference, however, the group with higher TKD mutant level had higher relapsed rate (6/8 and 3/6, respectively) (31).

Figure 3. Comparing sequence of sample 76 cloned into vector pUC19
Table 4: Mutant level of TKD mutations
| No. | % of 114 bp fragment uncut product | Mutant level (%) |
|
9
|
45.61
|
10.77
|
|
36
|
71.25
|
26.08
|
|
40
|
48.32
|
12.01
|
|
76
|
79.60
|
32.78
|
|
83
|
79.16
|
32.41
|
|
105
|
71.23
|
26.07
|
Conclusions
Two diagnosis procedures to detect FLT3 mutations in FMS-like tyrosine kinase 3 (FLT3) gene was successfully established and optimized. Applying these procedures to preliminary screen 156 Vietnamese AML patients, 14 FLT3-ITD (9.0%) and 6 FLT3-TKD (3.8%) mutations were found. ITDs ranged from 1 to 3 in number, with length between 20 to 90 bp, occurred at level from 4.6% to 63.7% and did not change the open reading frame. TKD mutations had mutant levels varied from 10% to 30% and sequences replaced G for T in codon 835, resulting in an Asp to Tyr change (D835Y). In the near future, we might apply the procedure in larger sample size or in cooperation with other clinical facilities to have more representative statistical data on AML gene mutations.
Acknowledgements
The research was conducted at Key Laboratory of Enzyme and Protein Technology, VNU University of Science. We would like to thanks Vietnam National Institute of Hematology and Blood Transfusion (NIHBT) for sample collection.



Comments
Virgilio replied on Permalink
Add new comment | Vietnam Journal of Science
Hi! This is my first comment here so I just wanted
to give a quick shout out and tell you I really enjoy reading
your articles. Can you recommend any other blogs/websites/forums that deal
with the same topics? Many thanks!
Also visit my web page - cleaning services ct
Add new comment