Yueng Hsing Ho, and Habibah A. Wahab*
Background
Type 2 insulin-resistant diabetes mellitus (T2DM) accounts for 90–95% of all diabetes. This heterogeneous disorder afflicts an estimated 6% of the adult population in Western society (1). By March 2013, World Health Organisation (WHO) reported approximately 347 million of diabetic patients worldwide. WHO also project that diabetes will be the seventh leading cause of death by year 2030 and more than 80% of diabetic death occurred in low and middle income countries. According to Malaysia National Health and Morbidity Survey (NHMS, 2011), the prevalence of diabetes in Malaysia are 15.2% which responsible for more than 2.6 million of Malaysian adults.
Current therapies for T2DM rely mainly on several approaches intended to reduce the hyperglycaemia itself and these therapies not only having limited efficacy, limited tolerability and also brought significant mechanism-based side effects (2).
Tyrosine-protein phosphatase non-receptor type 1 (PTP1B) is an enzyme localised at the cytoplasmic face of the endoplasmic reticulum and it is found in protein-tyrosine phosphatase (PTP) family which encoded by the PTPN1 gene in human body. PTP1B mainly function as a negative regulator in both insulin and leptin signalling pathways. PTP1B is the key regulator of insulin-receptor (IR) activity that acts at the insulin receptor and at downstream signalling components, such as insulin receptor substrate, IRS1 (3). Figure 1 illustrated the important sites and residues on PTP1B protein when interacting with its inhibitor.
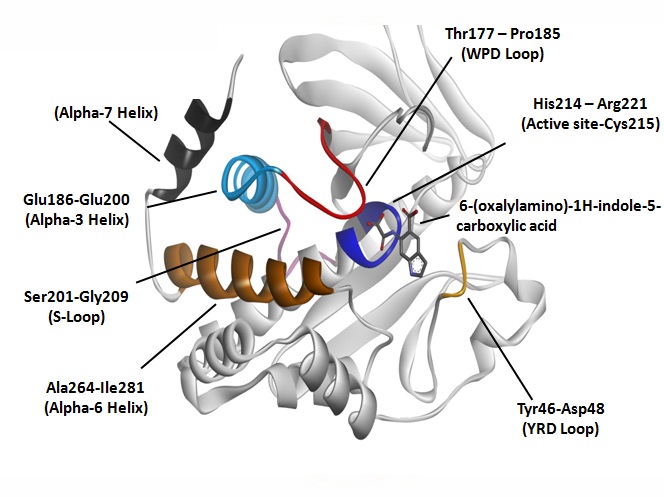
Fig. 1. Interaction between PTP1B protein and its known inhibitor 6-(oxalyamino)-1H-indole-5-carboxylic acid with labelled important sites and residues.
Both insulin and leptin resistance are the hallmark in common for T2DM and obesity as insulin is responsible for glucose uptake and lipogenesis while leptin is responsible for food intake and peripheral energy expenditure (4). More and more experimental evidences had shown that both insulin and leptin resistance are associated with the increase in activity or expression of PTP1B. In vivo, the PTP1B knock-outs mice displayed lower circulating insulin, glucose, and triglyceride levels, increased phosphorylation of the IR in liver and muscle after insulin injection, and improved insulin sensitivity in glucose and insulin tolerance tests (5), (6), (7). Here, we can conclude that inhibition of PTP1B could be the solution, not only to, T2DM but also weight gain or obesity triggered by T2DM onset.
Experimental and result
In this work, we are intended to find potential PTP1B inhibitors from Malaysian local plants using in silico approaches.First, the Protein Data Bank (PDB) structure 1C83 was selected as the macromolecule for the screening process. 1C83, consists of 298 residues, is a X-ray diffracted crystal structure of PTP1B with reasonable resolution of 1.80Å and related to another six similar PDB structures. Further verification and comparison can be easily done with its homologous structures which available in the PDB.
Besides that, 1C83 is a bounded protein structure with its inhibitor 6-(oxalylamino)-1H-indole-5-carboxylic acid (PDB identifier: OAI). Control docking was done using 1C83 against OAI with 62 X 62 X 62 Å grid points at 0.375 Å spacing and centered on potential binding site 44.746, 13.645, 2.842. The root-mean-square deviation (RMSD) of the control docking is 0.52Å with free energy of binding (FEB) of -10.11 kcal mol-1. Then, virtual screening was done using 1C83 against NADI database. A total of 3000 natural compounds from Malaysia local plants were screened against 1C83 and a few compounds were shortlisted based on their favorable FEB and interactions with 1C83. Figure 2 illustrated the top hit compounds from Piper retrofractum, Momordica charantia, Stemona tuberose, Garcinia mangostana and Cymbopogon citrates with FEB varied from -10.03 to -9.53 kcal mol-1. All the shortlisted compounds are attributed to some of the essential interactions between residues Tyr46, Val49, Lys120, Asp181, Phe182, Cys215, Ser216, Ile219, Gly220 and Arg221.
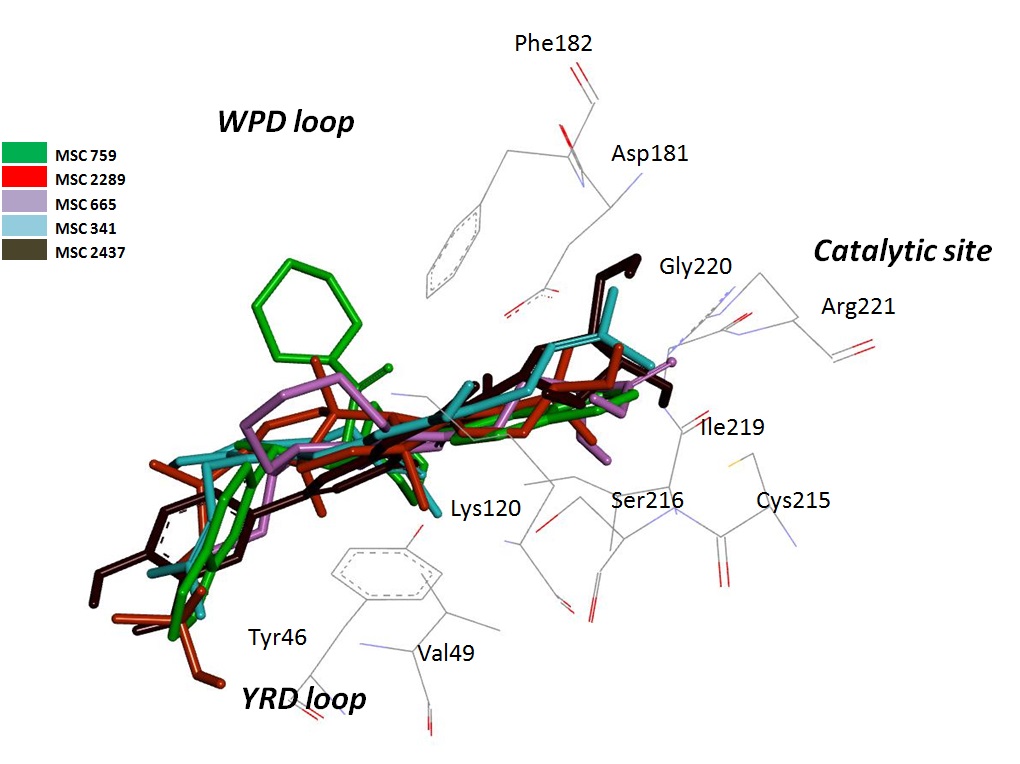
Fig. 2. Interaction between PTP1B protein and top hit compounds.
All docking procedure including virtual screening process were done using Autodock Tools versions 1.5.6 and AutoDock 4.2 while interaction analyses and visualization were done using Discovery Studio 2.5, Accelrys Software Inc..
Conclusions
These findings provided useful understanding on the inhibition interaction between PTP1B and its potential inhibitors. These findings also provided potential inhibitor candidates for future T2DM studies and drug development.



Add new comment